A basic overview of multiple data analysis
Compiled: 二月 14, 2023
Source:vignettes/multibatch_basic_tutorial.Rmd
multibatch_basic_tutorial.RmdOverview
This tutorial demonstrates how to use SRTpipeline (>=0.1.0) to analyze multiple spatially-resolved transcriptomics (SRT) data. We emphases the workflow based on the SRTProject object in the SRTpipeline package. This tutorial will cover the following tasks, which we believe will be common for many SRT data analyses:
- Normalization
- Feature selection
- Spatial clustering
- Data integration
- Batch-corrected gene expression
- DEG analysis
- Spatial trajectory inference
- Visualization
First, we load SRTpipeline package.
library(SRTpipeline)
set.seed(2023) # set a random seed for reproducibilityDataset
For this tutorial, we will introduce how to create a SRTProject object with multiple SRT samples using SRTpipeline that includes an introduction to common analytical workflows for multiple data batches. Here, we will be taking spatial transcriptomics dataset for mouse liver as an example. There are eight tissue slices with 450~700 spots and 8500~9500 genes that were sequenced on the ST platform. Our preprocessed data can be downloaded here, and the raw data can be found here.
Next, we download the data to the current working path for the followed analysis by the following command:
seuList <- readRDS("seuList_liver8.RDS")Pre-processing workflow
Prepare SRTProject
Then load to R.
n_sample <- length(seuList)
library(Seurat)
## create count matrix list
cntList <- list()
## create spatial coordinate matrix
coordList <- list()
## create metadata list
metadataList <- list()
for (r in 1:n_sample) {
# r <- 1
message("r = ", r)
seu <- seuList[[r]]
sp_count <- seu@assays$RNA@counts
meta.data <- seu@meta.data
row.names(meta.data) <- colnames(sp_count)
cntList[[r]] <- sp_count
metadataList[[r]] <- meta.data
coordList[[r]] <- cbind(row = seu$row, col = seu$col)
}
names(cntList) <- paste0("Liver", 1:n_sample) # Note each component in count matrix list has a name.
## create meta data for each data batches.
sampleMetadata <- data.frame(species = rep("Mouse", n_sample), tissues = rep("Liver", n_sample))
row.names(sampleMetadata) <- names(cntList)
## Name of this project
projectName <- "Liver8"Create a SRTProject object
We start creating SRTProject object. Filter out the genes less than 20 spots with nonzero read counts and the spots less than 20 genes with nonzero read counts.
SRTProj <- CreateSRTProject(cntList, coordList, projectName = projectName, metadataList, sampleMetadata,
min.spots = 20, min.genes = 20)
SRTProj## class: SRTProject
## outputPath: F:\Research paper\IntegrateDRcluster\AnalysisCode\SRTpipeline\vignettes\Liver8
## h5filePath: F:\Research paper\IntegrateDRcluster\AnalysisCode\SRTpipeline\vignettes\Liver8/Liver8.h5
## ---------Datasets basic information-----------------
## samples(8): Liver1 Liver2 ... Liver7 Liver8
## sampleColData names(3): species tissues NumOfSpots
## cellMetaData names(13): orig.ident nCount_RNA ... sample_id batch
## numberOfSpots(8): 675 684 ... 629 590
## ---------Downstream analyses information-----------------
## Low-dimensional embeddings(0):
## Inferred cluster labels: No
## Embedding for plotting(0):Normalizing the data
After removing unwanted cells and genes from the dataset, the next step is to normalize the data. To save RAM memory, normalized values are stored in disk as a h5file.
SRTProj <- normalizeSRT(SRTProj, normalization.method = "LogNormalize")Feature selection
We next select a subset of genes that exhibit high spot-to-spot variation in the dataset (i.e, they are highly expressed in some spots, and lowly expressed in others). It has been found that focusing on these genes in downstream analysis helps to highlight biological signal in single-cell datasets here.
Then we choose variable features. The default number of variable features is 2,000, and users can change it using argument nfeatures. The default type is highly variable genes (HVGs), but users can use spatially variable genes by seting type='SVGs', then SPARK-X will be used to choose SVGs.
SRTProj <- selectVariableFeatures(SRTProj, nfeatures = 2000, type = "HVGs", method = "vst")
SRTProj## class: SRTProject
## outputPath: F:\Research paper\IntegrateDRcluster\AnalysisCode\SRTpipeline\vignettes\Liver8
## h5filePath: F:\Research paper\IntegrateDRcluster\AnalysisCode\SRTpipeline\vignettes\Liver8/Liver8.h5
## ---------Datasets basic information-----------------
## samples(8): Liver1 Liver2 ... Liver7 Liver8
## sampleColData names(3): species tissues NumOfSpots
## cellMetaData names(13): orig.ident nCount_RNA ... sample_id batch
## numberOfSpots(8): 675 684 ... 629 590
## ---------Downstream analyses information-----------------
## Variable features: 2000
## Low-dimensional embeddings(0):
## Inferred cluster labels: No
## Embedding for plotting(0):Calculate the adjcence matrix
## Obtain adjacency matrix
SRTProj <- AddAdj(SRTProj, platform = "ST")Dimension reduction
PCA is the most popular dimension reduction technique in single cell RNA sequencing (scRNA-seq) data and SRT data because of its simplicity, computational efficiency, and relatively comparable performance. SRTpipeline provided three versions of PCA: standard PCA (PCA), approximated PCA (APCA), and weighted PCA (WPCA) by the function AddPCA with default version as APCA for fastest computation. The 15-dimensional PCs are extracted by default, and users can use their own values.
After running AddPCA, we would see the output of SRTProj includes PCA in the Low-dimensional embeddings field.
## APCA
SRTProj <- AddPCA(SRTProj, n_comp = 15)
# accurate PCA SRTProj <- AddPCA(SRTProj, Method='PCA')
# weighted PCA SRTProj <- AddPCA(SRTProj, Method='WPCA')
SRTProj## class: SRTProject
## outputPath: F:\Research paper\IntegrateDRcluster\AnalysisCode\SRTpipeline\vignettes\Liver8
## h5filePath: F:\Research paper\IntegrateDRcluster\AnalysisCode\SRTpipeline\vignettes\Liver8/Liver8.h5
## ---------Datasets basic information-----------------
## samples(8): Liver1 Liver2 ... Liver7 Liver8
## sampleColData names(3): species tissues NumOfSpots
## cellMetaData names(13): orig.ident nCount_RNA ... sample_id batch
## numberOfSpots(8): 675 684 ... 629 590
## ---------Downstream analyses information-----------------
## Variable features: 2000
## Low-dimensional embeddings(1): PCA
## Inferred cluster labels: No
## Embedding for plotting(0):Data clustering and integration analysis using iSC.MEB model
iSC.MEB model achieves joint clustering and alignment by integration analysis for multiple samples. Some SRT clustering methods use Markov random field to model clusters of spots. These approaches work extremely well when the spots have strong spatial dependence. iSC.MEB extracts the micro-environment related embeddings and aligned embeddings. The micro-environment related and aligned embeddings are saved in the slot reductions$microEnv.iSC.MEB and reductions$aligned.iSC.MEB, and the spatial clusters are saved in the slot clusters.
SRTProj <- Integrate_iSCMEB(SRTProj, K = 6, reduction = "PCA")## fitting ...
##
|
| | 0%
|
|=================================== | 50%
|
|======================================================================| 100%
## Finish variable initialization
## K = 6, iter = 2, loglik= -39897.400172, dloglik=0.999981
## K = 6, iter = 3, loglik= -36353.590917, dloglik=0.088823
## K = 6, iter = 4, loglik= -35341.453489, dloglik=0.027841
## K = 6, iter = 5, loglik= -34875.854831, dloglik=0.013174
## K = 6, iter = 6, loglik= -34573.520344, dloglik=0.008669
## K = 6, iter = 7, loglik= -34346.587951, dloglik=0.006564
## K = 6, iter = 8, loglik= -34159.814691, dloglik=0.005438
## K = 6, iter = 9, loglik= -33996.841377, dloglik=0.004771
## K = 6, iter = 10, loglik= -33848.331818, dloglik=0.004368
## K = 6, iter = 11, loglik= -33710.517314, dloglik=0.004072
## K = 6, iter = 12, loglik= -33582.502133, dloglik=0.003797
## K = 6, iter = 13, loglik= -33465.382937, dloglik=0.003488
## K = 6, iter = 14, loglik= -33361.001693, dloglik=0.003119
## K = 6, iter = 15, loglik= -33268.844201, dloglik=0.002762
## K = 6, iter = 16, loglik= -33186.458205, dloglik=0.002476
## K = 6, iter = 17, loglik= -33111.504371, dloglik=0.002259
## K = 6, iter = 18, loglik= -33042.335693, dloglik=0.002089
## K = 6, iter = 19, loglik= -32977.845549, dloglik=0.001952
## K = 6, iter = 20, loglik= -32917.269207, dloglik=0.001837
## K = 6, iter = 21, loglik= -32860.061645, dloglik=0.001738
## K = 6, iter = 22, loglik= -32805.830764, dloglik=0.001650
## K = 6, iter = 23, loglik= -32754.294333, dloglik=0.001571
## K = 6, iter = 24, loglik= -32705.245790, dloglik=0.001497
## K = 6, iter = 25, loglik= -32658.526356, dloglik=0.001428
SRTProj## class: SRTProject
## outputPath: F:\Research paper\IntegrateDRcluster\AnalysisCode\SRTpipeline\vignettes\Liver8
## h5filePath: F:\Research paper\IntegrateDRcluster\AnalysisCode\SRTpipeline\vignettes\Liver8/Liver8.h5
## ---------Datasets basic information-----------------
## samples(8): Liver1 Liver2 ... Liver7 Liver8
## sampleColData names(3): species tissues NumOfSpots
## cellMetaData names(13): orig.ident nCount_RNA ... sample_id batch
## numberOfSpots(8): 675 684 ... 629 590
## ---------Downstream analyses information-----------------
## Variable features: 2000
## Low-dimensional embeddings(3): PCA microEnv.iSC.MEB aligned.iSC.MEB
## Inferred cluster labels: Yes
## Embedding for plotting(0):Visualization
SRT embeddings
To check the performance of integration, we visualize the inferred clusters and data batches on the two-dimensional tSNEs. First, we use AddTSNE() function to calculate the two-dimensional tSNEs based on the aligned.iSC.MEB embeddings.
SRTProj <- AddTSNE(SRTProj, n_comp = 2, reduction = "aligned.iSC.MEB")We visualized the inferred clusters and data batches. The tSNE plot showed the domain clusters were well segregated and data batches were well mixed.
cols_cluster <- chooseColors(n_colors = 6)
cols_batch <- chooseColors(palettes_name = "Light 13", n_colors = 8)
p_tsne2_cluster <- EmbedPlot(SRTProj, item = "cluster", plotEmbeddings = "tSNE", cols = cols_cluster,
legend.position = "right", pt_size = 0.2)
p_tsne2_batch <- EmbedPlot(SRTProj, item = "batch", plotEmbeddings = "tSNE", cols = cols_batch,
legend.position = "right", pt_size = 0.2)
drawFigs(list(p_tsne2_cluster, p_tsne2_batch), layout.dim = c(1, 2), legend.position = "right")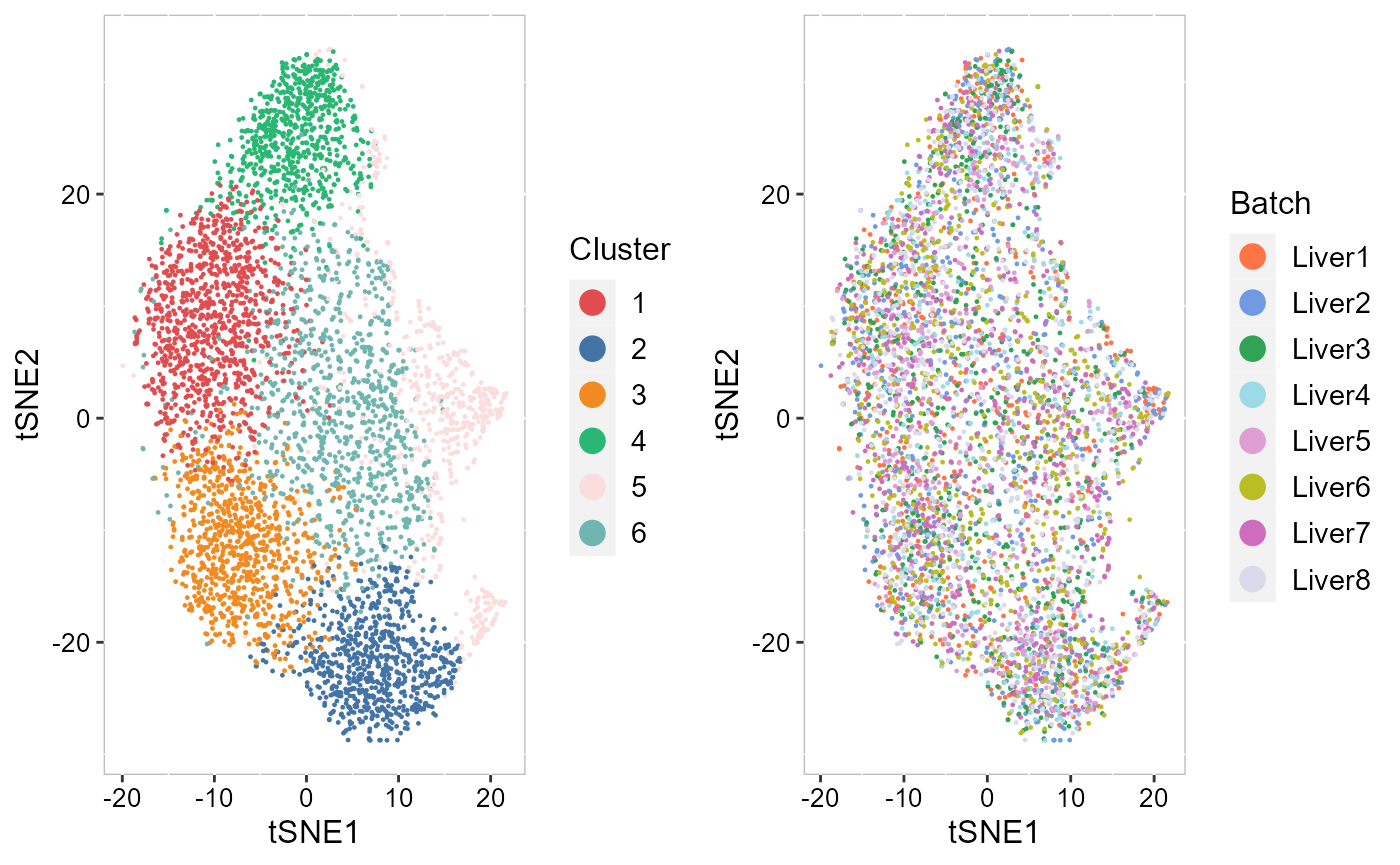
Spatial heatmap to show the clustering performance
Except for the embedding plots, SRTpipeline also provides a variaty of visualization functions. First, we visualize the spatial distribution of cluster labels that shows the tissue structure for all data batches.
p12 <- EachClusterSpaHeatMap(SRTProj, cols = cols_cluster, legend.position = "bottom", base_size = 12,
pt_size = 2, layout.dim = c(2, 4), nrow.legend = 1)
p12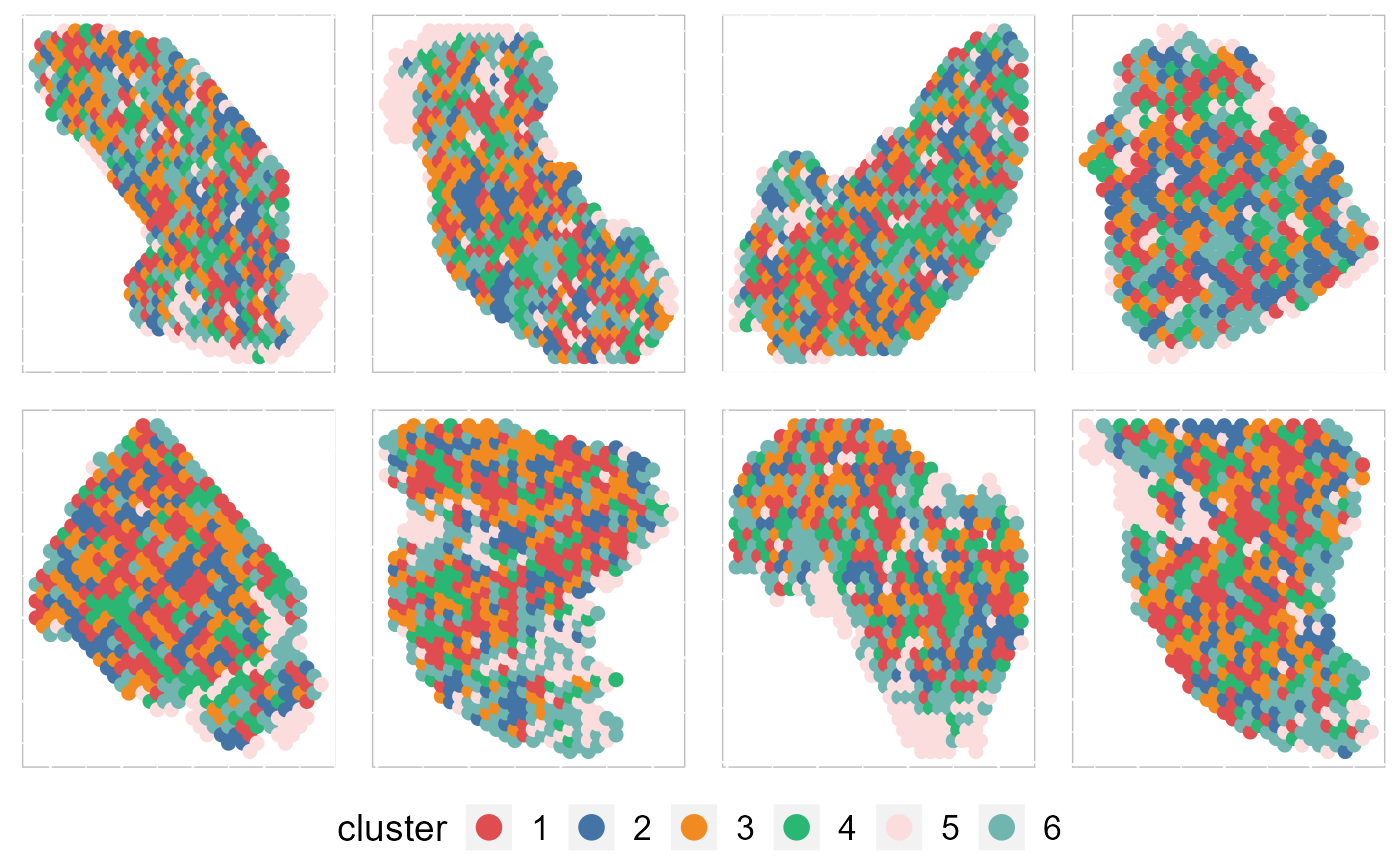 By setting
By setting combine =FALSE, this function will return a list of ggplot2 objects, thus user can revise each plot. In addtion, we can also plot some of data batches that are interested.
pList <- EachClusterSpaHeatMap(SRTProj, cols = cols_cluster, legend.position = "bottom", base_size = 12,
pt_size = 0.5, layout.dim = c(2, 4), nrow.legend = 1, combine = FALSE)
EachClusterSpaHeatMap(SRTProj, batch = 1:4, title_name = "iSC.MEB: ", cols = cols_cluster, legend.position = "right",
base_size = 12, pt_size = 2, layout.dim = c(2, 2))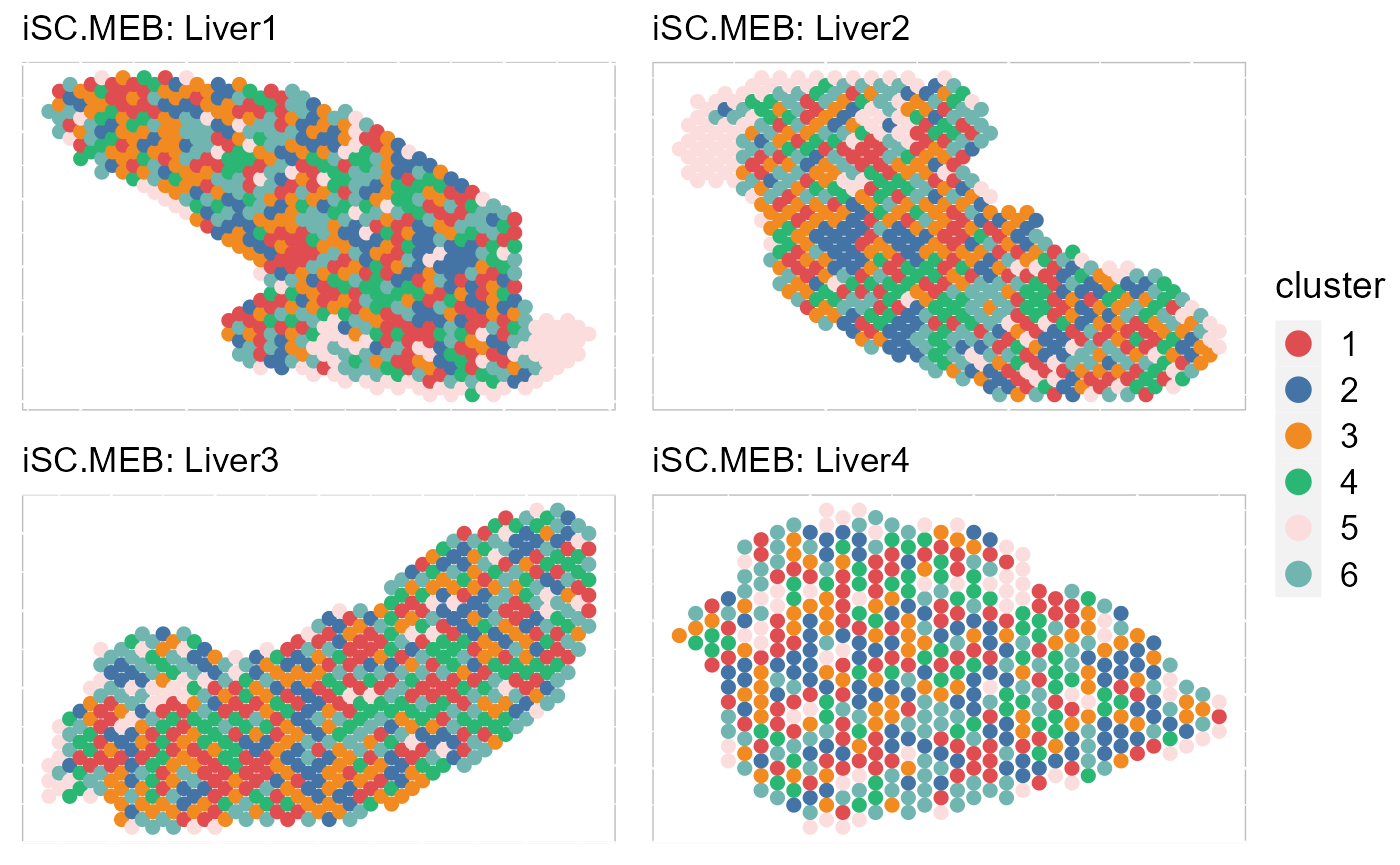
RGB plot to show the embedding performance
Next, we summarized the inferred embeddings for biological effects between spatial domain types (the slot reductions) using three components from either tSNE or UMAP and visualized the resulting tSNE/UMAP components with red/green/blue (RGB) colors in the RGB plot.
The resulting RGB plots from iSC.MEB showed the structure organization of the mouse liver tissue, and iSC.MEB provided fast transitions across portal vein and central vein domains.
SRTProj <- AddTSNE(SRTProj, n_comp = 3, reduction = "aligned.iSC.MEB")
p_tsne3 <- EachRGBSpaHeatMap(SRTProj, plot_type = "tSNE", pt_size = 1, title_name = "", layout.dim = c(2,
4))
p_tsne3 To run UMAP in SRTpipeline we use the
To run UMAP in SRTpipeline we use the AddUMAP() function. Frist, we evaluate the two-dimensional UMAPs.
SRTProj <- AddUMAP(SRTProj, n_comp = 2, reduction = "aligned.iSC.MEB")
p_umap2_cluster <- EmbedPlot(SRTProj, item = "cluster", plotEmbeddings = "UMAP", cols = cols_cluster,
legend.position = "bottom")
p_umap2_batch <- EmbedPlot(SRTProj, item = "batch", plotEmbeddings = "UMAP", cols = cols_batch,
legend.position = "bottom")
drawFigs(list(p_umap2_cluster, p_umap2_batch), layout.dim = c(1, 2), legend.position = "bottom")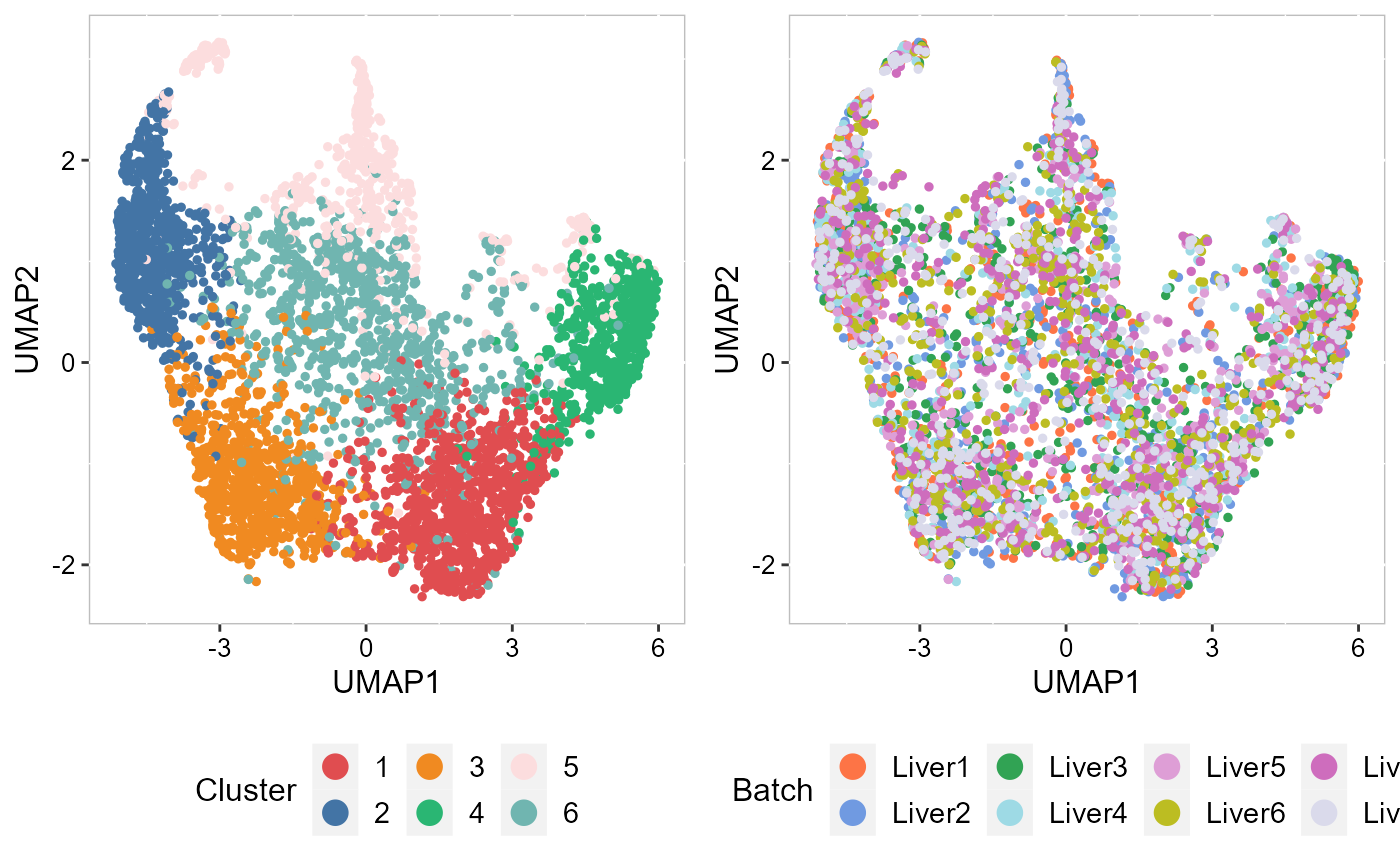 Then, we evaluate the three-dimensional UMAPs.
Then, we evaluate the three-dimensional UMAPs.
SRTProj <- AddUMAP(SRTProj, n_comp = 3, reduction = "aligned.iSC.MEB")
p_umap3 <- EachRGBSpaHeatMap(SRTProj, plot_type = "UMAP", layout.dim = c(2, 4), pt_size = 1.2, title_name = "UMAP: ")
p_umap3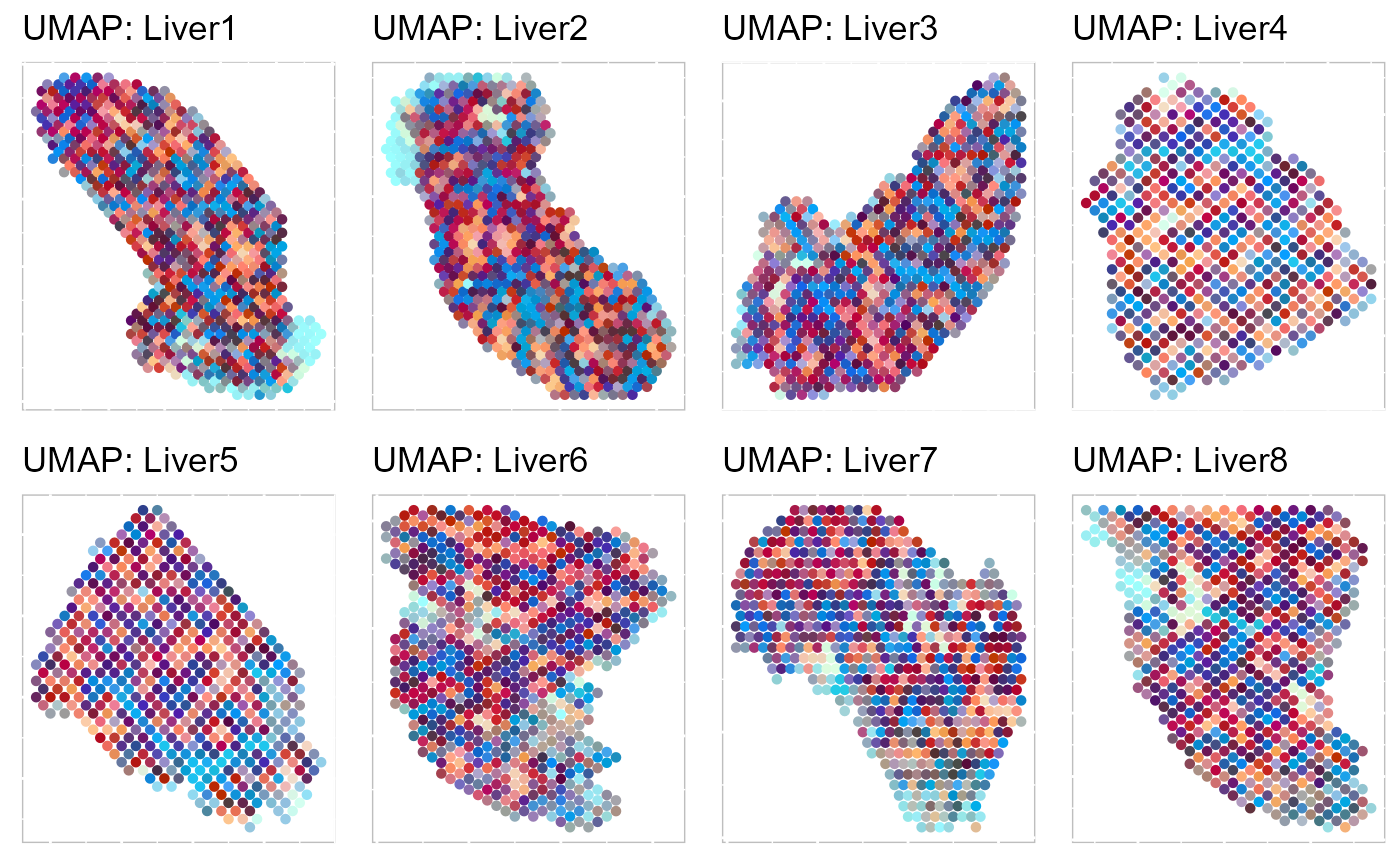
To save the plot, we can use write_fig() function.
write_fig(p_umap3, filename = "iSCMEB_p_umap3.png", width = 14, height = 11)Moreover, we can visualize the microenvironment effects on the spatial coordinates. Then, we evaluate the three-dimensional UMAPs based on microEnv.iSC.MEB.
# save the previous calculated UMAP3
umap3_cluster <- SRTProj@plotEmbeddings$UMAP3
SRTProj <- AddUMAP(SRTProj, n_comp = 3, reduction = "microEnv.iSC.MEB")
p_umap3_micro <- EachRGBSpaHeatMap(SRTProj, plot_type = "UMAP", layout.dim = c(2, 4), pt_size = 1.2,
title_name = "mircoEnv: ")
p_umap3_micro
Cell-by-cell heatmap to show the relation of spatial domains
We plotted the heatmap of Pearson’s correlation coefcients of the estimated embeddings among the detected domains shows the good separation of the estimated embeddings across domains and the high correlations between domains with similarity.
p_cc <- CCHeatMap(SRTProj, reduction = "aligned.iSC.MEB", grp_color = cols_cluster)
p_cc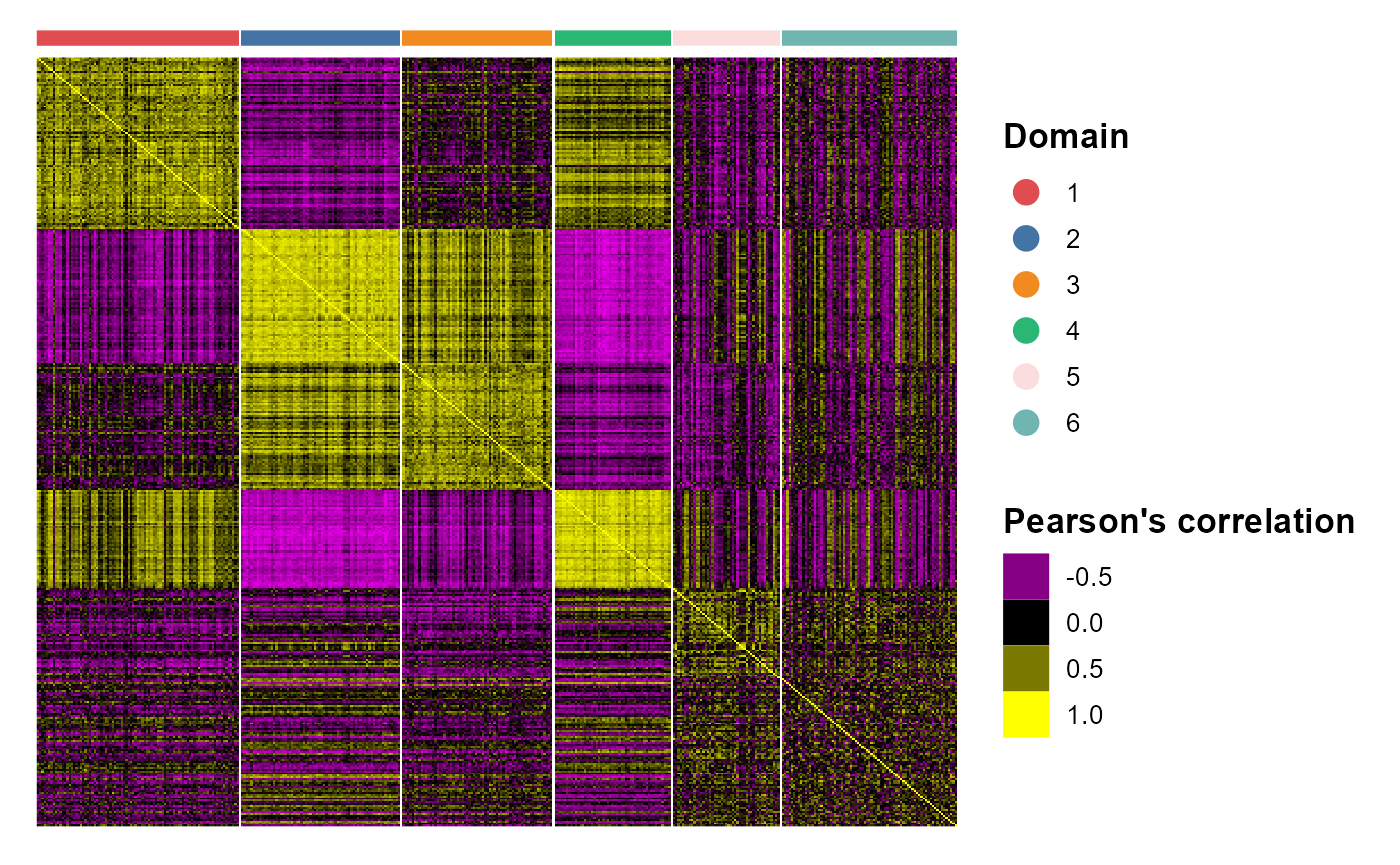 After adding the quantities for data visualization, the
After adding the quantities for data visualization, the SRTProject object will have more information in the downstream analyses information. Now, we print this SRTProject object to check it. We observed two components added in the slot plotEmbeddings (Embeddings for plotting): tSNE, tSNE3, UMAP and UMAP3.
SRTProj## class: SRTProject
## outputPath: F:\Research paper\IntegrateDRcluster\AnalysisCode\SRTpipeline\vignettes\Liver8
## h5filePath: F:\Research paper\IntegrateDRcluster\AnalysisCode\SRTpipeline\vignettes\Liver8/Liver8.h5
## ---------Datasets basic information-----------------
## samples(8): Liver1 Liver2 ... Liver7 Liver8
## sampleColData names(3): species tissues NumOfSpots
## cellMetaData names(13): orig.ident nCount_RNA ... sample_id batch
## numberOfSpots(8): 675 684 ... 629 590
## ---------Downstream analyses information-----------------
## Variable features: 2000
## Low-dimensional embeddings(3): PCA microEnv.iSC.MEB aligned.iSC.MEB
## Inferred cluster labels: Yes
## Embedding for plotting(4): tSNE tSNE3 UMAP UMAP3Combined DEG analysis
After fitting the integration model iSC.MEB, we could obatin the batch corrected log-normalized gene expression data by using function getIntegratedData().
speInt <- getIntegratedData(SRTProj, Method = "iSC.MEB", species = "Mouse")After obtain the spatial cluster labels using a clustering model (i.e., iSC.MEB), we can perform differentially expression analysis. First, we can detect the DE genes using function FindDEGs for only one cluster, i.e., cluster 3.
## DataFrame with 1 row and 5 columns
## p_val avg_log2FC pct.1 pct.2 p_val_adj
## <numeric> <numeric> <numeric> <numeric> <numeric>
## A1bg 3.04919e-42 0.252504 0.979 0.877 6.09838e-39We perform differential expression analysis for all clusters by using FindAllDEGs() function, then the DE genes’ information is saved in a data.frame object dat_degs.
dat_degs <- FindAllDEGs(speInt)
dat_degs## DataFrame with 137 rows and 7 columns
## p_val avg_log2FC pct.1 pct.2 p_val_adj cluster
## <numeric> <numeric> <numeric> <numeric> <numeric> <factor>
## Cyp2f2 1.30039e-98 -0.250081 0.987 0.973 2.60078e-95 1
## Cyp3a44 1.63897e-63 0.314311 0.928 0.779 3.27793e-60 1
## Sds 1.77854e-48 -0.355872 0.802 0.827 3.55708e-45 1
## Aldh1b1 5.89245e-22 -0.264130 0.672 0.727 1.17849e-18 1
## Cyp2e1 1.15125e-274 -0.423678 0.993 0.998 2.30251e-271 2
## ... ... ... ... ... ... ...
## Rnf13 1.20494e-16 -0.250187 0.820 0.943 2.40988e-13 5
## Dpt 1.41514e-14 0.295305 0.639 0.545 2.83028e-11 5
## Vim 9.67010e-09 0.326605 0.671 0.647 1.93402e-05 5
## Col1a1 6.84510e-06 0.302746 0.599 0.572 1.36902e-02 5
## Col1a2 7.85131e-03 0.327749 0.601 0.612 1.00000e+00 5
## gene
## <character>
## Cyp2f2 Cyp2f2
## Cyp3a44 Cyp3a44
## Sds Sds
## Aldh1b1 Aldh1b1
## Cyp2e1 Cyp2e1
## ... ...
## Rnf13 Rnf13
## Dpt Dpt
## Vim Vim
## Col1a1 Col1a1
## Col1a2 Col1a2We identify the significant DE genes by two criteria: (a) adjustd p-value less than 0.01 and (b) average log fold change greater than 0.15.
degs_sig <- subset(dat_degs, p_val_adj < 0.01 & avg_log2FC > 0.15)
degs_sig## DataFrame with 69 rows and 7 columns
## p_val avg_log2FC pct.1 pct.2 p_val_adj cluster
## <numeric> <numeric> <numeric> <numeric> <numeric> <factor>
## Cyp3a44 1.63897e-63 0.314311 0.928 0.779 3.27793e-60 1
## Cyp2f2.1 3.97931e-223 0.387100 1.000 0.972 7.95862e-220 2
## Sds.1 1.08390e-213 0.659946 0.984 0.791 2.16781e-210 2
## Hal 4.06491e-196 0.427699 0.999 0.933 8.12982e-193 2
## Ctsc 2.60875e-186 0.508951 0.993 0.848 5.21749e-183 2
## ... ... ... ... ... ... ...
## Slc13a3 9.57091e-26 0.291170 0.659 0.559 1.91418e-22 4
## Gsn 2.01150e-36 0.567004 0.748 0.611 4.02300e-33 5
## Col3a1 2.37755e-27 0.491134 0.771 0.699 4.75510e-24 5
## Dpt 1.41514e-14 0.295305 0.639 0.545 2.83028e-11 5
## Vim 9.67010e-09 0.326605 0.671 0.647 1.93402e-05 5
## gene
## <character>
## Cyp3a44 Cyp3a44
## Cyp2f2.1 Cyp2f2
## Sds.1 Sds
## Hal Hal
## Ctsc Ctsc
## ... ...
## Slc13a3 Slc13a3
## Gsn Gsn
## Col3a1 Col3a1
## Dpt Dpt
## Vim VimIn the following, we perform gene set enrichment analysis for the DE genes of each spatial Domain identified by iSC.MEB model using R package gprofiler2.
library(gprofiler2)
termList <- list()
for (k in 1:6) {
# k <- 1
if (sum(degs_sig$cluster == k) > 0) {
cat("k = ", k, "\n")
dat_degs_sub <- subset(degs_sig, cluster == k)
que1 <- dat_degs_sub$gene
gostres <- gost(query = que1, organism = "hsapiens", correction_method = "fdr")
termList[[k]] <- gostres
}
}## k = 1
## k = 2
## k = 3
## k = 4
## k = 5
head(termList[[1]]$result)## NULLTo understand the functions of the identified spatial domains by iSC.MEB model, we compare the top significant biological process (BP) pathways in GO database for the DE genes from Domain 1 and 4. Here, we only show the significant BP pathways and users can explore the other databases such as KEGG and HPA.
## Most commonly used databases
source_set <- c("GO:BP", "GO:CC", "GO:MF", "KEGG", "HPA")
cols <- c("steelblue3", "goldenrod", "brown3", "#f98866", "#CE6DBD")
## Here, we show GO:BP
source1 <- "GO:BP"
ss <- which(source_set == source1)
ntop = 5
names(cols) <- source_set
pList_enrich <- list()
for (ii in 1:5) {
## ii <- 3
message("ii=", ii)
gostres2 <- termList[[ii]]
if (!is.null(gostres2)) {
dat1 <- subset(gostres2$result, term_size < 500)
dat1 <- get_top_pathway(dat1, ntop = ntop, source_set = source1)
dat1 <- dat1[complete.cases(dat1), ]
dat1$nlog10P <- -log10(dat1$p_value)
pList_enrich[[ii]] <- barPlot_enrich(dat1[order(dat1$nlog10P), ], source = "source", "term_name",
"nlog10P", cols = cols[source_set[ss]], base_size = 14) + ylab("-log10(p-adj)") + xlab("Biological terms") +
ggtitle(paste0("Domain", ii))
}
}
drawFigs(pList_enrich[c(3, 4)], layout.dim = c(2, 1), common.legend = T, align = "hv")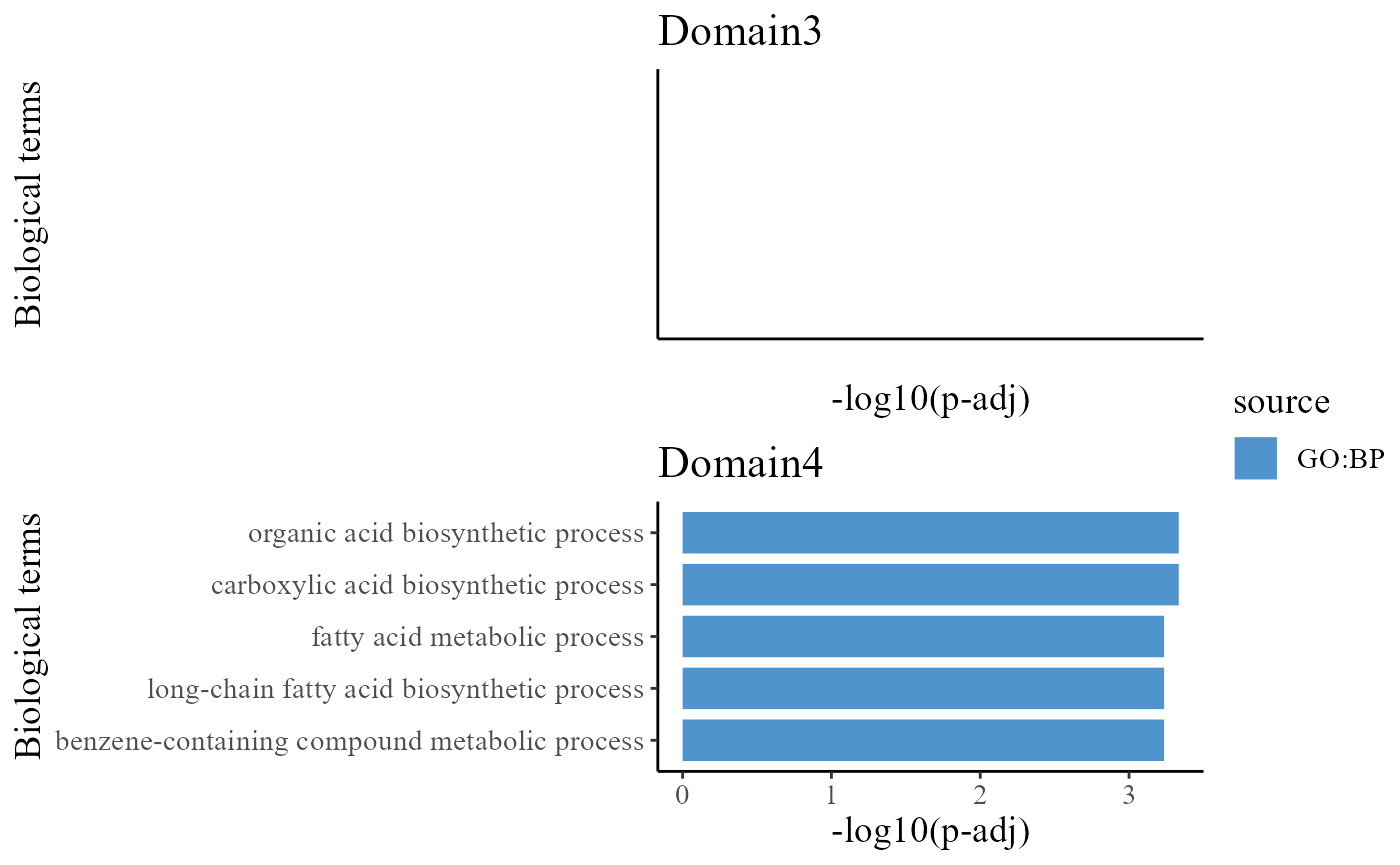
We take out the top DE genes for each cluster for visualization.
library(dplyr)
n <- 5
dat_degs %>%
as.data.frame %>%
group_by(cluster) %>%
top_n(n = n, wt = avg_log2FC) -> topGene
topGene## # A tibble: 22 x 7
## # Groups: cluster [5]
## p_val avg_log2FC pct.1 pct.2 p_val_adj cluster gene
## <dbl> <dbl> <dbl> <dbl> <dbl> <fct> <chr>
## 1 1.30e- 98 -0.250 0.987 0.973 2.60e- 95 1 Cyp2f2
## 2 1.64e- 63 0.314 0.928 0.779 3.28e- 60 1 Cyp3a44
## 3 1.78e- 48 -0.356 0.802 0.827 3.56e- 45 1 Sds
## 4 5.89e- 22 -0.264 0.672 0.727 1.18e- 18 1 Aldh1b1
## 5 1.08e-213 0.660 0.984 0.791 2.17e-210 2 Sds
## 6 2.61e-186 0.509 0.993 0.848 5.22e-183 2 Ctsc
## 7 1.24e-166 0.629 0.925 0.677 2.48e-163 2 Aldh1b1
## 8 2.60e-107 0.475 0.919 0.728 5.21e-104 2 Etnppl
## 9 1.27e- 72 0.524 0.8 0.599 2.54e- 69 2 Spp1
## 10 3.05e- 42 0.253 0.979 0.877 6.10e- 39 3 A1bg
## # ... with 12 more rowsWe visualize the DE genes for each cluster group by gene-by-cell heatmap using the GCHeatMap() function.
p1 <- GCHeatMap(speInt, features = topGene$gene, grp_color = cols_cluster, y_text_size = 12)
p1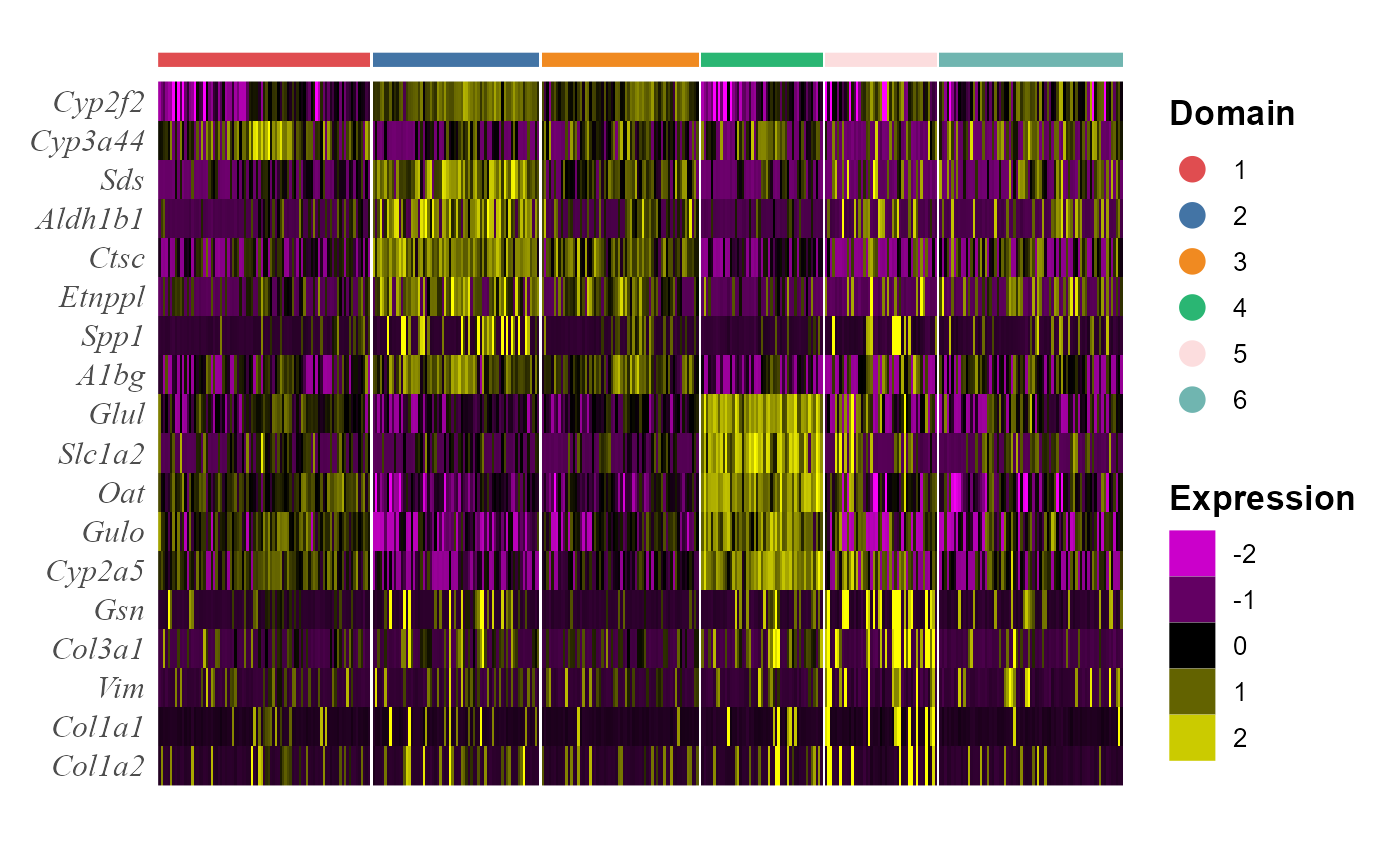
Trajectory inference
Next, we performed trajectory inference using the aligned embeddings and domain labels estimated by iSC.MEB model.
speInt <- AddTrajectory(speInt, reduction = "aligned.iSC.MEB")
p1 <- EmbedPlot(speInt, reduction = "aligned.iSC.MEB", colour_by = "PT")
p2 <- EmbedPlot(speInt, reduction = "tSNE", colour_by = "PT")
drawFigs(list(p1, p2), layout.dim = c(1, 2), common.legend = TRUE, legend.position = "right")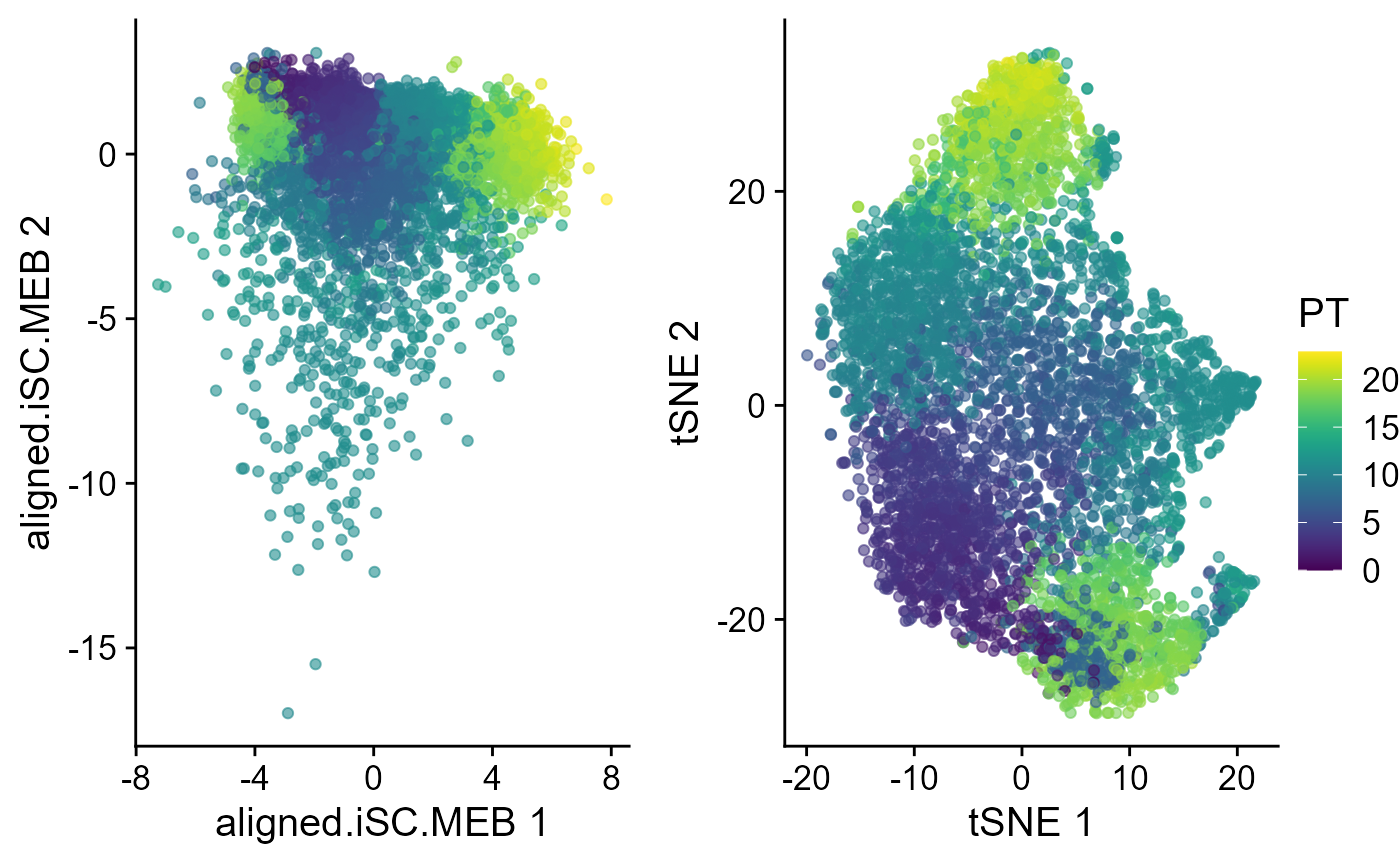
Visualize the inferred pseudotime on the spatial coordinates for each data batch.
p_spa <- EachEmbedPlot(speInt, reduction = "Coord", colour_by = "PT", layout.dim = c(2, 4))
p_spa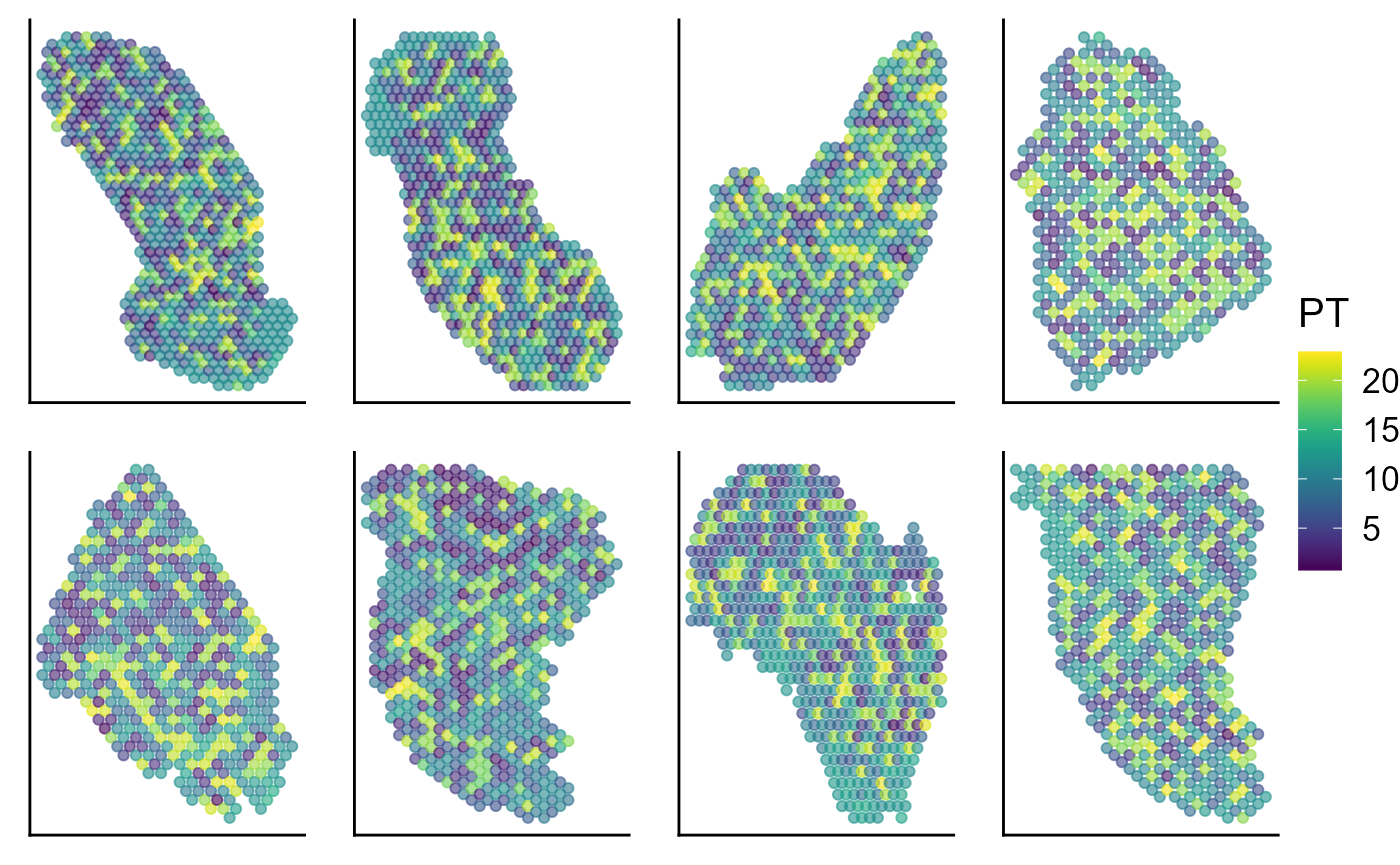
# save(SRTProj, file=paste0(SRTProj@projectMetadata$outputPath,'/SRTProj.rds'))
# load('F:/Research
# paper/IntegrateDRcluster/AnalysisCode/SRTpipeline/vignettes/Liver8/SRTProj.rds')Other downstream analyses
Session Info
## R version 4.1.2 (2021-11-01)
## Platform: x86_64-w64-mingw32/x64 (64-bit)
## Running under: Windows 10 x64 (build 22621)
##
## Matrix products: default
##
## locale:
## [1] LC_COLLATE=Chinese (Simplified)_China.936
## [2] LC_CTYPE=Chinese (Simplified)_China.936
## [3] LC_MONETARY=Chinese (Simplified)_China.936
## [4] LC_NUMERIC=C
## [5] LC_TIME=Chinese (Simplified)_China.936
##
## attached base packages:
## [1] parallel stats4 stats graphics grDevices utils datasets
## [8] methods base
##
## other attached packages:
## [1] scater_1.25.1 scuttle_1.4.0
## [3] slingshot_2.2.0 TrajectoryUtils_1.2.0
## [5] princurve_2.1.6 gprofiler2_0.2.1
## [7] bigalgebra_1.1.0 bigmemory_4.5.36
## [9] SpatialExperiment_1.4.0 SingleCellExperiment_1.16.0
## [11] SummarizedExperiment_1.24.0 Biobase_2.54.0
## [13] GenomicRanges_1.46.1 GenomeInfoDb_1.30.1
## [15] IRanges_2.28.0 MatrixGenerics_1.6.0
## [17] matrixStats_0.62.0 dplyr_1.0.9
## [19] colorspace_2.0-3 iSC.MEB_1.0.1
## [21] ggplot2_3.3.6 gtools_3.9.2.2
## [23] irlba_2.3.5 DR.SC_3.1
## [25] spatstat.geom_2.4-0 spatstat.data_3.0-0
## [27] Matrix_1.4-0 hdf5r_1.3.5
## [29] ff_4.0.7 bit_4.0.4
## [31] S4Vectors_0.32.3 BiocGenerics_0.40.0
## [33] rhdf5_2.38.0 sp_1.5-0
## [35] SeuratObject_4.1.0 Seurat_4.1.1
## [37] SRTpipeline_0.1.1
##
## loaded via a namespace (and not attached):
## [1] scattermore_0.8 ggthemes_4.2.4
## [3] R.methodsS3_1.8.1 GiRaF_1.0.1
## [5] ragg_1.2.2 tidyr_1.2.0
## [7] bit64_4.0.5 knitr_1.37
## [9] DelayedArray_0.20.0 R.utils_2.11.0
## [11] data.table_1.14.2 rpart_4.1.16
## [13] RCurl_1.98-1.6 generics_0.1.2
## [15] ScaledMatrix_1.2.0 cowplot_1.1.1
## [17] RANN_2.6.1 future_1.26.1
## [19] httpuv_1.6.5 assertthat_0.2.1
## [21] viridis_0.6.2 xfun_0.29
## [23] jquerylib_0.1.4 evaluate_0.15
## [25] promises_1.2.0.1 fansi_1.0.3
## [27] igraph_1.3.5 DBI_1.1.2
## [29] htmlwidgets_1.5.4 purrr_0.3.4
## [31] ellipsis_0.3.2 RSpectra_0.16-1
## [33] ggpubr_0.4.0 backports_1.4.1
## [35] deldir_1.0-6 sparseMatrixStats_1.6.0
## [37] vctrs_0.4.1 ROCR_1.0-11
## [39] abind_1.4-5 cachem_1.0.6
## [41] withr_2.5.0 PRECAST_1.4
## [43] progressr_0.10.1 sctransform_0.3.3
## [45] mclust_5.4.10 goftest_1.2-3
## [47] cluster_2.1.2 lazyeval_0.2.2
## [49] crayon_1.5.1 edgeR_3.36.0
## [51] pkgconfig_2.0.3 labeling_0.4.2
## [53] nlme_3.1-155 vipor_0.4.5
## [55] rlang_1.0.2 globals_0.15.0
## [57] lifecycle_1.0.1 miniUI_0.1.1.1
## [59] bigmemory.sri_0.1.3 rsvd_1.0.5
## [61] rprojroot_2.0.3 polyclip_1.10-0
## [63] lmtest_0.9-40 carData_3.0-5
## [65] Rhdf5lib_1.16.0 zoo_1.8-10
## [67] beeswarm_0.4.0 ggridges_0.5.3
## [69] rjson_0.2.21 png_0.1-7
## [71] viridisLite_0.4.0 bitops_1.0-7
## [73] R.oo_1.24.0 KernSmooth_2.23-20
## [75] rhdf5filters_1.6.0 DelayedMatrixStats_1.16.0
## [77] stringr_1.4.0 parallelly_1.32.0
## [79] spatstat.random_2.2-0 rstatix_0.7.0
## [81] ggsignif_0.6.3 beachmat_2.10.0
## [83] scales_1.2.0 memoise_2.0.1
## [85] magrittr_2.0.3 plyr_1.8.7
## [87] ica_1.0-2 zlibbioc_1.40.0
## [89] compiler_4.1.2 dqrng_0.3.0
## [91] RColorBrewer_1.1-3 fitdistrplus_1.1-8
## [93] cli_3.2.0 XVector_0.34.0
## [95] listenv_0.8.0 patchwork_1.1.1
## [97] pbapply_1.5-0 formatR_1.11
## [99] MASS_7.3-55 mgcv_1.8-39
## [101] tidyselect_1.1.2 stringi_1.7.6
## [103] textshaping_0.3.6 highr_0.9
## [105] yaml_2.3.6 BiocSingular_1.10.0
## [107] locfit_1.5-9.4 ggrepel_0.9.1
## [109] grid_4.1.2 sass_0.4.1
## [111] tools_4.1.2 future.apply_1.9.0
## [113] rstudioapi_0.13 gridExtra_2.3
## [115] farver_2.1.0 Rtsne_0.16
## [117] DropletUtils_1.14.2 digest_0.6.29
## [119] rgeos_0.5-9 shiny_1.7.1
## [121] Rcpp_1.0.10 car_3.0-12
## [123] broom_0.7.12 later_1.3.0
## [125] RcppAnnoy_0.0.19 httr_1.4.3
## [127] fs_1.5.2 tensor_1.5
## [129] reticulate_1.25 splines_4.1.2
## [131] uwot_0.1.11 spatstat.utils_3.0-1
## [133] pkgdown_2.0.6 plotly_4.10.0
## [135] systemfonts_1.0.4 xtable_1.8-4
## [137] jsonlite_1.8.0 R6_2.5.1
## [139] pillar_1.7.0 htmltools_0.5.2
## [141] mime_0.12 glue_1.6.2
## [143] fastmap_1.1.0 BiocParallel_1.28.3
## [145] BiocNeighbors_1.12.0 codetools_0.2-18
## [147] utf8_1.2.2 lattice_0.20-45
## [149] bslib_0.3.1 spatstat.sparse_2.1-1
## [151] tibble_3.1.7 ggbeeswarm_0.6.0
## [153] leiden_0.4.2 magick_2.7.3
## [155] survival_3.2-13 limma_3.50.1
## [157] CompQuadForm_1.4.3 rmarkdown_2.11
## [159] desc_1.4.0 munsell_0.5.0
## [161] GenomeInfoDbData_1.2.7 HDF5Array_1.22.1
## [163] reshape2_1.4.4 gtable_0.3.0
## [165] spatstat.core_2.4-4